UTGB Toolkit 1.5.8
:: DESCRIPTION
UTGB Toolkit (University of Tokyo Genome Browser) is an open-source software for developing personalized genome browsers that work in web browsers.
::DEVELOPER
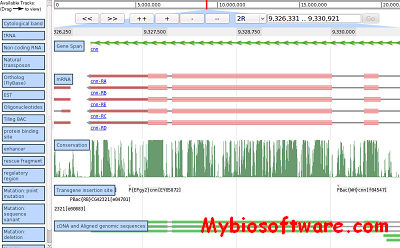
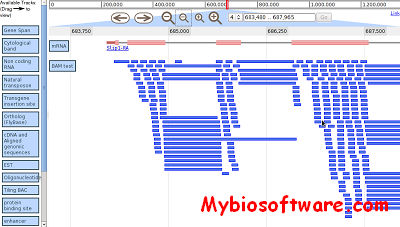
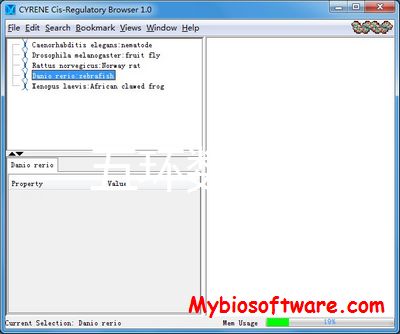
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / Mac OsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2009 Aug 1;25(15):1856-61. Epub 2009 Jun 3.
UTGB toolkit for personalized genome browsers.
Saito TL, Yoshimura J, Sasaki S, Ahsan B, Sasaki A, Kuroshu R, Morishita S.