SecretomeP 2.0
:: DESCRIPTION
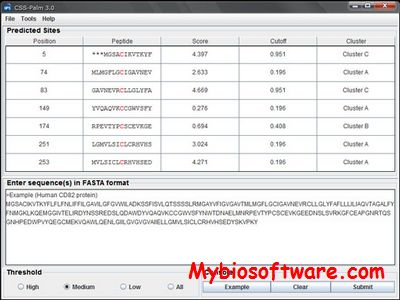
SecretomeP server produces ab initio predictions of non-classical i.e. not signal peptide triggered protein secretion. The method queries a large number of other feature prediction servers to obtain information on various post-translational and localizational aspects of the protein, which are integrated into the final secretion prediction.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Feature based prediction of non-classical and leaderless protein secretion
J. Dyrløv Bendtsen, L. Juhl Jensen, N. Blom, G. von Heijne and S. Brunak
Protein Eng. Des. Sel., 17(4):349-356, 2004