POPGENE 1.32
:: DESCRIPTION
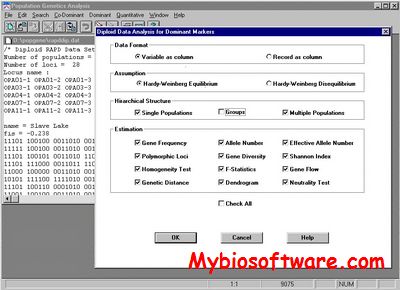
POPGENE is a user-friendly computer freeware for the analysis of genetic variation among and within populations using co-dominant and dominant markers. This package provides the Windows graphical user interface that makes population genetics analysis more accessible for the casual computer user and more convenient for the experienced computer user. Simple menus and dialog box selections enable you to perform complex analyses and produce scientifically sound statistics, thereby assisting you to adequately analyze population genetic structure using the target markers.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION