Flapjack 1.22.04.21
:: DESCRIPTION
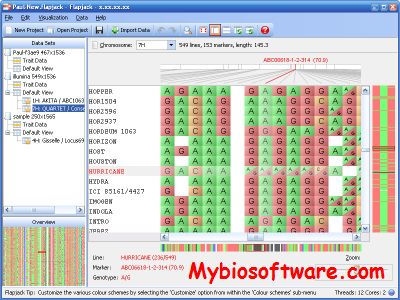
Flapjack is a new software tool for graphical genotyping and haplotype visualization which is required that can routinely handle the large data volumes generated by high throughput SNP and comparable genotyping technologies. Flapjack is a new visualization tool to facilitate analysis of these data types. Its visualizations are rendered in real-time allowing for rapid navigation and comparisons between lines, markers and chromosomes.
::DEVELOPER
Information & Computational Sciences, The James Hutton Institute
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOSX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Iain Milne etc.
Flapjack – Graphical Genotype Visualization
Bioinformatics (2010)doi : 10.1093/bioinformatics/btq580