ViTraM 2.0
:: DESCRIPTION
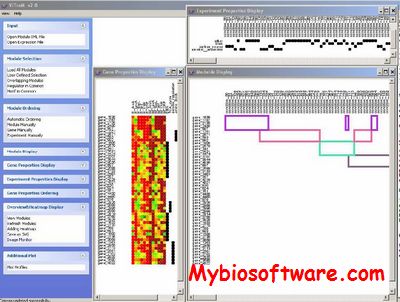
ViTraM (Visualization of Transcriptional Modules) is a flexible software platform for visualizing overlapping transcriptional modules in an intuitive way. By visualizing not only the genes and the experiments in which the genes are co-expressed, but also additional properties of the modules such as the regulators and regulatory motifs that are responsible for the observed co-expression, ViTraM can assist in the biological analysis and interpretation of the output of module detection tools.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / MacOsX
- Java
:: MORE INFORMATION
Citation
H. Sun, K. Lemmens, T. Van den Buckle, K. Engelen, B. De Moor, K. Marchal.
“ViTraM: Visualization of Transcriptional Modules” (2009).
Bioinformatics, 25(18):2450-2451;