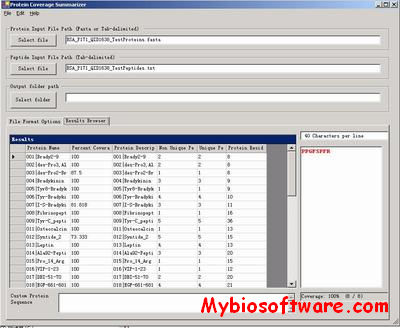
HSEpred
:: DESCRIPTION
HSEpred is a web server to predict the HSE(Half-Sphere Exposure) measures and infer residue contact numbers using the predicted HSE values, based on a well-prepared non-homologous protein structure dataset.
::DEVELOPER
Akutsu Laboratory (Laboratory of Mathematical Bioinformatics)
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- Web Server
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Bioinformatics. 2008 Jul 1;24(13):1489-97. doi: 10.1093/bioinformatics/btn222.
HSEpred: predict half-sphere exposure from protein sequences.
Song J, Tan H, Takemoto K, Akutsu T.