CoPlot 6.45
:: DESCRIPTION
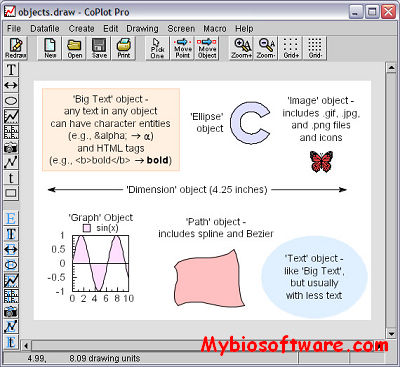
CoPlot is an incredibly versatile program for making publication-quality 2D and 3D scientific graphs (which plot data and equations), maps, and technical drawings. Every aspect of CoPlot has been designed around one basic goal: to make a program that is so versatile that scientists and engineers can easily get exactly what they want. CoPlot includes CoStat for data handling and statistics.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Mac OsX / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION