HypoTrace
:: DESCRIPTION
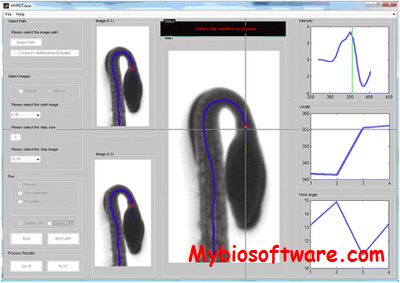
HypoTrace takes time series of image files as input and output growth rate as well as hook angle measurements for Hypocotyls. The software comes with a build-in Graphical User Interface, which allows the software to be run under three modes: 1. Automatic mode with no user intervention, good for high quality images; 2. Semi-Auto mode with a little bit user intervention, good for fair quality images; 3. Manual mode, work for all images including really bad quality images. The three modes are designed to effectively detect the cotyledons bifurcation point under all conditions.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOsX
- Matlab
:: DOWNLOAD
:: MORE INFORMATION
Citation
Wang L., Uilecan I. V., Assadi A. H., Kozmik C. A., Spalding E. P.,
HypoTrace: Image Analysis Software for Measuring Hypocotyl Growth and Shape Demonstrated on Arabidopsis Seedlings Undergoing Photomorphogenesis,
Plant Physiol. 2009, 149(4), 1632-1637.