CleanCollapse 1.0.5
:: DESCRIPTION
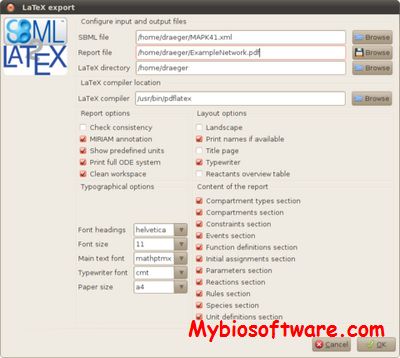
CleanCollapse is a relatively user-friendly Windows program that accepts an alignment of sequences, and optionally can remove rare sporadic polymorphisms that may represent analytical artifacts, and also reduce redundancy by “collapsing” identical sequences into a single sequence. The program provides a record of these manipulation, and some rudimentary analysis (e.g. proportion of synonymous and nonsynonymous sporadic changes) that may assist interpretation
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Before you download software you need to read disclaimer.