BioPAX2SBML
:: DESCRIPTION
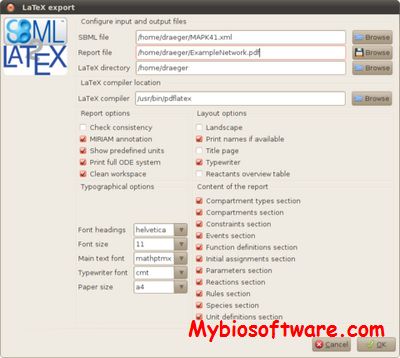
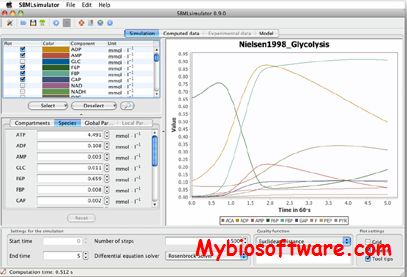
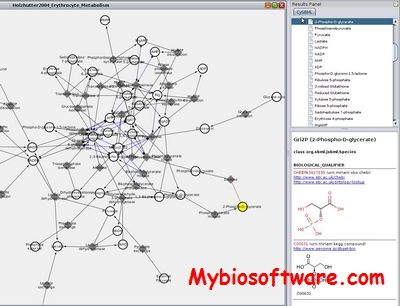
BioPAX2SBML is a webtool. It is capable of properly translating BioPAX formats (Level 2 and 3) into SBML format (Level 3 Version 1 with qual) including reactions and relations in one model.
::DEVELOPER
the Interfaculty Institute for Biomedical Informatics (IBMI)
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Büchel F, Wrzodek C, Mittag F, Dräger A, Eichner J, Rodriguez N, Le Novère N, and Zell A.
Qualitative translation of relations from BioPAX to SBML.
Bioinformatics (2012) 28 (20): 2648-2653.