SCA
:: DESCRIPTION
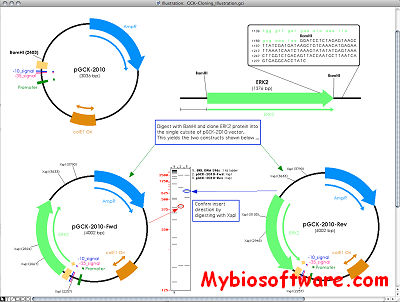
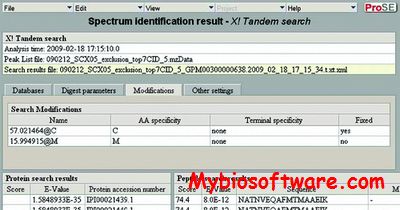
The SCA (sequence composition analysis) takes a set of sequences in fasta format and generates a 2-dimensional crossed graph displaying the value of a given formula on the x-axis against the year of the sequences on the y-axis. The year is taken from the header line of each source sequence (starting with “>”). Additionally the points may be colored depending of the host the sequence is taken from.
::DEVELOPER
Institute of Bioinformatics WWU Muenster
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows / MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION