iVUN 1.2
:: DESCRIPTION
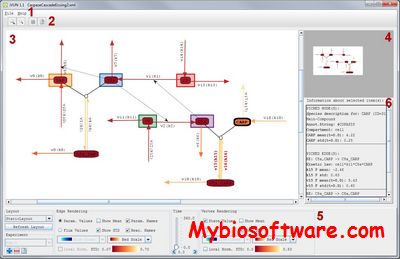
iVUN is a visualization toolbox which supports uncertainty-aware analysis of static and dynamic attributes of biochemical reaction networks
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
C. Vehlow, J. Hasenauer, A. Kramer, J. Heinrich, N. Radde, F. Allgoewer, and D. Weiskopf.
Uncertainty-aware visual analysis of biochemical reaction networks.
In Proceedings of IEEE Symposium on Biological Data Visualization(Biovis), pages 91–98, 2012.