SplitsTree 6.1.10
:: DESCRIPTION
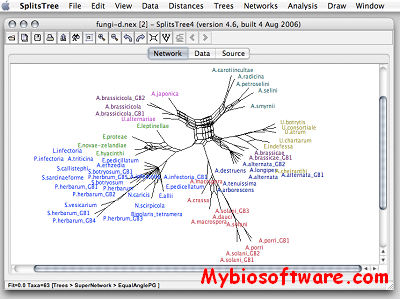
SplitsTree uses the split decomposition method to analyse and visualize distance data, e.g. data derived from biosequences. SplitsTree4 is the leading application for computing unrooted phylogenetic networks from molecular sequence data. Given an alignment of sequences, a distance matrix or a set of trees, the program will compute a phylogenetic tree or network using methods such as split decomposition, neighbor-net, consensus network, super networks methods or methods for computing hybridization or simple recombination networks.
::DEVELOPER
the Algorithms in Bioinformatics lab.
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / Mac OsX
:: DOWNLOAD
:: MORE INFORMATION
Citation
D. H. Huson and D. Bryant,
Application of Phylogenetic Networks in Evolutionary Studies
Mol. Biol. Evol., 23(2):254-267, 2006.