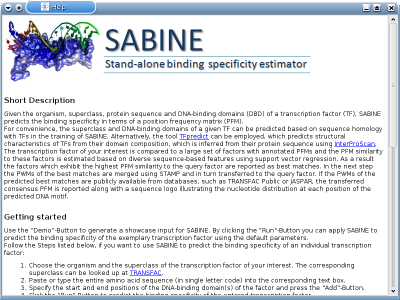
TRAP 3.05
:: DESCRIPTION
TRAP (Transcription factor Affinity Prediction) calculates the affinity of transcription factors for DNA sequences on the basis of a biophysical model. This method has proven to be useful for several applications, including for determining the putative target genes of a given factor. This protocol covers two other applications: (i) determining which transcription factors have the highest affinity in a set of sequences (illustrated with chromatin immunoprecipitation–sequencing (ChIP-seq) peaks), and (ii) finding which factor is the most affected by a regulatory single-nucleotide polymorphism.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- C++ compiler
- R Package
:: DOWNLOAD
:: MORE INFORMATION
Citation
Morgane Thomas-Chollier, Andrew Hufton, Matthias Heinig, Sean O’Keeffe, Nassim El Masri, Helge G Roider, Thomas Manke and Martin Vingron.
Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs.
Nature Protocols, 3;6(12):1860-9. (2011)