PhyloTrack 2.1
:: DESCRIPTION
PhyloTrack is a JavaScript–based software tool that integrates the D3.js library for data visualization with the JBrowse tool for genome browser representation.
::DEVELOPER
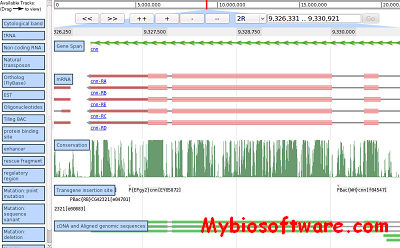
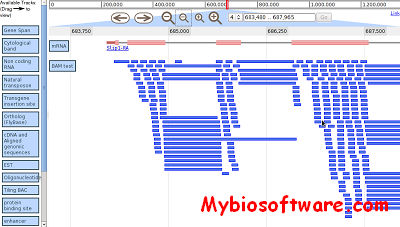
:: SCREENSHOTS
::REQUIREMENTS
- Linux
- Perl
- JavaScript
:: DOWNLOAD
:: MORE INFORMATION
Citation
PhyTB: Phylogenetic tree visualisation and sample positioning for M. tuberculosis.
Benavente ED, Coll F, Furnham N, McNerney R, Glynn JR, Campino S, Pain A, Mohareb FR, Clark TG.
BMC Bioinformatics. 2015 May 13;16:155. doi: 10.1186/s12859-015-0603-3.