CGView 1.0
:: DESCRIPTION
CGView (Circular Genome Viewer) is a Java application and library for generating high-quality, zoomable maps of circular genomes.Its primary purpose is to serve as a component of sequence annotation pipelines, as a means of generating visual output suitable for the web. Feature information and rendering options are supplied to the program using an XML file, a tab delimited file, or an NCBI ptt file. CGView converts the input into a graphical map (PNG, JPG, or Scalable Vector Graphics format), complete with labels, a title, legends, and footnotes. In addition to the default full view map, the program can generate a series of hyperlinked maps showing expanded views. The linked maps can be explored using any web browser, allowing rapid genome browsing, and facilitating data sharing. The feature labels in maps can be hyperlinked to external resources, allowing CGView maps to be integrated with existing web site content or databases.
::DEVELOPER
Paul Stothard
:: SCREENSHOTS
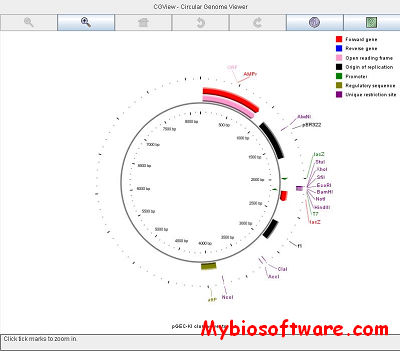
:: REQUIREMENTS
- Windows / Mac / Linux
- Java
:: DOWNLOAD
 CGView
CGView
:: MORE INFORMATION
Citation:
Stothard P, Wishart DS.
Circular genome visualization and exploration using CGView.
Bioinformatics 21:537-539.
 NO
NO