QMSIM 1.10
:: DESCRIPTION
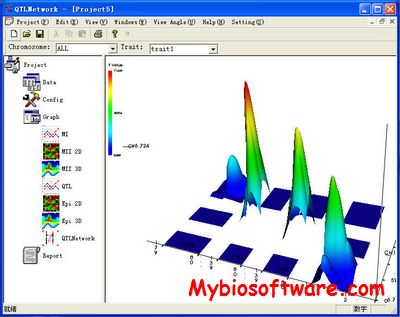
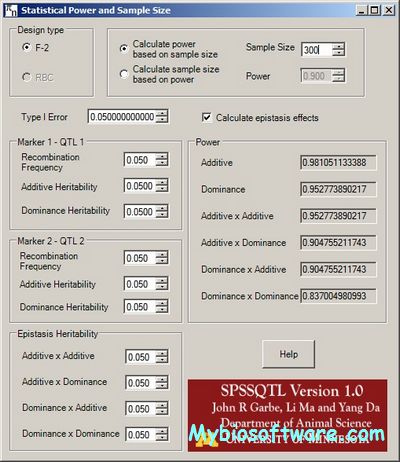
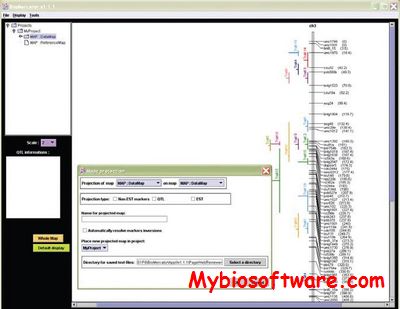
QMSim (Qtl and Marker SIMulator) was designed to simulate a wide range of genetic architectures and population structures in livestock. Large scale genotyping data and complex pedigrees can be efficiently simulated. QMSim is a family based simulator, which can also take into account predefined evolutionary features, such as LD, mutation, bottlenecks and expansions. The simulation is basically carried out in two steps: In the first step, a historical population is simulated to establish mutation-drift equilibrium and, in the second step, recent population structures are generated, which can be complex. QMSim allows for a wide range of parameters to be incorporated in the simulation models in order to produce appropriate simulated data.
::DEVELOPER
Mehdi Sargolzaei , Flavio Schenkel
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Sargolzaei, M. and F. S. Schenkel. 2009.
QMSim: a large-scale genome simulator for livestock.
Bioinformatics, 25: 680-681. doi:10.1093 /bioinformatics/btp045