StarGenetics
:: DESCRIPTION
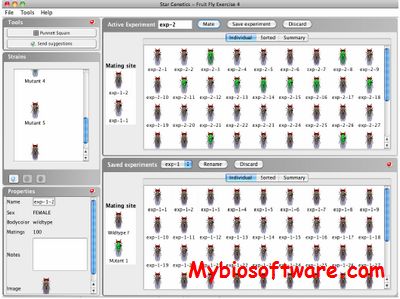
StarGenetics is a Mendelian genetics cross simulator. StarGenetics allows students to simulate mating experiments between organisms that are genetically different across a range of traits to analyze the nature of the traits in question. Its goal is to teach students about genetic experimental design and genetic concepts.
:: DEVELOPER
The STAR program at MIT
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows / MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION