MAPDISTO 1.8.1
:: DESCRIPTION
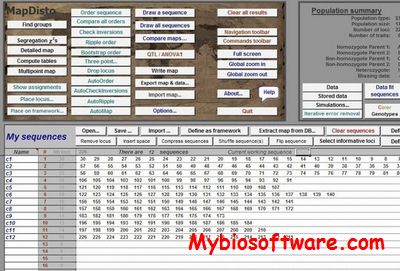
MAPDISTO is a program for mapping genetic markers in experimental segregating populations like backcross, doubled haploids, single-seed descent. Its specificity is to propose recombination fraction estimates in case of segregation distortion. It can (1) compute and draw genetic maps easily and quickly through a graphical interface; (2) facilitate the analysis of marker data showing segregation distortion due to differential viability of gametes or zygotes.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / MacOsX
- Excel
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Lorieux M (2012)
MapDisto: fast and efficient computation of genetic linkage maps.
Molecular Breeding, August 2012, Volume 30, Issue 2, pp 1231-1235