CCT
:: DESCRIPTION
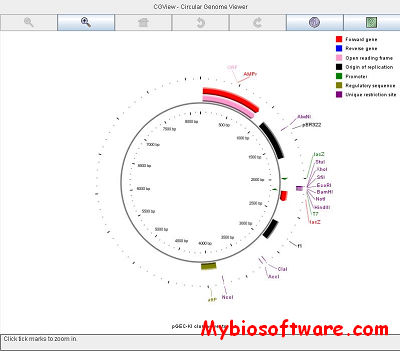
CCT is a package for visually comparing bacterial, plasmid, chloroplast, or mitochondrial sequences of interest to existing genomes or sequence collections. The comparisons are conducted using BLAST, and the BLAST results are presented in the form of graphical maps that can also show sequence features, gene and protein names, COG category assignments, and sequence composition characteristics. CCT can generate maps in a variety of sizes, including 400 Megapixel maps suitable for posters. Comparisons can be conducted within a particular species or genus, or all available genomes can be used. The entire map creation process, from downloading sequences to redrawing zoomed maps, can be completed easily using scripts included with the CCT. User-defined features or analysis results can be included on maps, and maps can be extensively customized.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Grant JR, Arantes AS, Stothard P (2012)
Comparing thousands of circular genomes using the CGView Comparison Tool.
BMC Genomics 13:202.