ThunderSTORM 1.3
:: DESCRIPTION
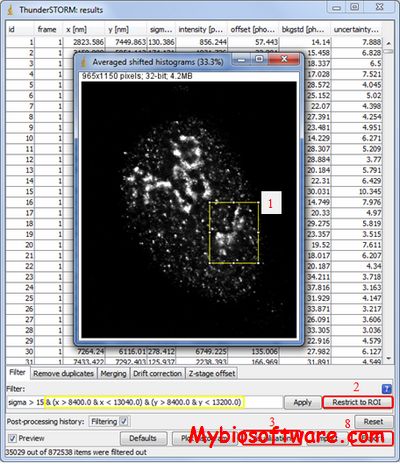
ThunderSTORM is an open-source, interactive and modular plug-in for ImageJ designed for automated processing, analysis and visualization of data acquired by single-molecule localization microscopy methods such as photo-activated localization microscopy and stochastic optical reconstruction microscopy.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
- ImageJ
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2014 Apr 25. pii: btu202. [Epub ahead of print]
ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging.
Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM.