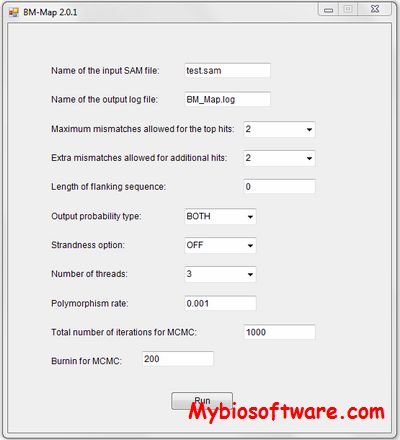
TreQ 0.1 beta
:: DESCRIPTION
TreQ is a read mapper for high-throughput DNA sequencing reads, in particular reads from 100 nt to hundreds of nucleotides, and for large edit distance between sequencing read and match in the reference genome.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- GCC
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2012 Sep 15;28(18):i325-i332. doi: 10.1093/bioinformatics/bts380.
Indel-tolerant read mapping with trinucleotide frequencies using cache-oblivious kd-trees.
Mahmud MP, Wiedenhoeft J, Schliep A.