UBViz 0.9R04
:: DESCRIPTION
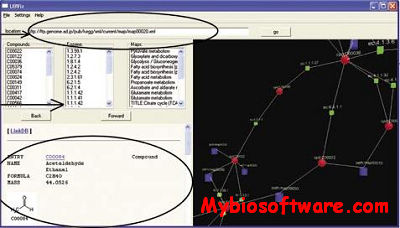
UBViz is a software tool designed to make biochemical pathways easier to understand for biologists. Users can visualize and interact with a biochemical pathway from KEGG’s website in 3-D space, and browse all related web pages within one window.
With UBViz’ built-in network functionalities, pathway data are obtained directly from KEGG’s website and automatically processed. As a result, users are guaranteed to view the newest version of the biochemical pathways. UBViz uses the popular XML format as its input, with very little modification UBViz can be easily adapted to view any database which stores pathway information in XML.
::DEVELOPER
Complex Computation Laboratory ,Iowa State University
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows /Mac OsX
:: DOWNLOAD
 Registration First ; UBViz
Registration First ; UBViz
:: MORE INFORMATION
Citation
UBViz: a software tool for exploring metabolic pathways in 3-D space
Song Li and Hui-Hsien Chou
BioTechniques Vol. 38, No. 4: pp 540-542 (Apr 2005)