Dizzy 1.11.4
:: DESCRIPTION
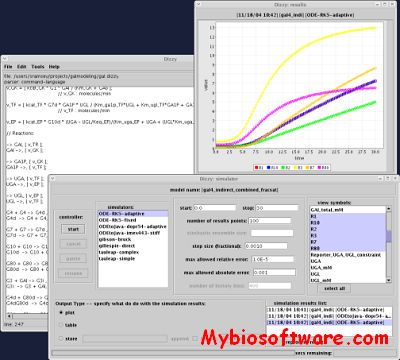
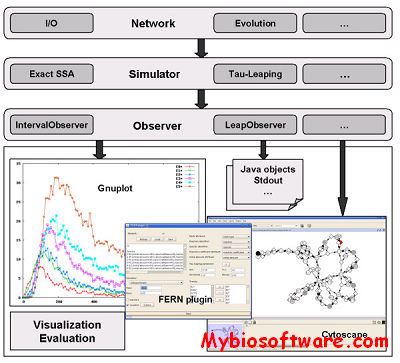
Dizzy is a chemical kinetics stochastic simulation software package written in Java. It provides a model definition environment and an implementation of the Gillespie, Gibson-Bruck, and Tau-Leap stochastic algorithms. Dizzy is capable of importing and exporting the SBML model definition language, as well as displaying models graphically using the Cytoscape software system.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux
- JAVA
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Ramsey S, Orrell D, Bolouri H. , “Dizzy: stochastic simulation of large-scale genetic regulatory networks.”
J Bioinform Comput Biol. 2005 Apr;3(2):415-36.