QSVanalyser 20121206
:: DESCRIPTION
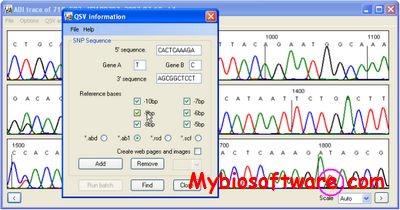
QSVanalyser is a program that allows rapid, user-controlled, batchwise analysis of DNA sequence traces for estimation of the relative proportions of two sequence variants (Quantitative Sequence Variants, QSVs).
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
- Microsoft .NET framework version 2.0
:: DOWNLOAD
:: MORE INFORMATION
Citation
Carr IM, Robinson JI, Dimitriou R, Markham AF, Morgan AW, Bonthron DT.
Inferring relative proportions of DNA variants from sequencing electropherograms
Bioinformatics 2009 December 25:3244-3250