MyMiner
:: DESCRIPTION
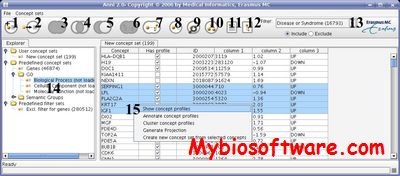
MyMiner is a free and user-friendly text annotation tool aimed to assist in carrying out the main biocuration tasks and to provide labelled data for the development of text mining systems.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
Bioinformatics. 2012 Sep 1;28(17):2285-7. Epub 2012 Jul 12.
MyMiner: a web application for computer-assisted biocuration and text annotation.
Salgado D, Krallinger M, Depaule M, Drula E, Tendulkar AV, Leitner F, Valencia A, Marcelle C.
 NO
NO