DiagAF 21.2.7
:: DESCRIPTION
DiagAF is a more accurate and efficient pre-alignment filter for sequence alignment. It can efficiently filter out candidates that contain errors greater than the edit distance threshold during read mapping.
::DEVELOPER
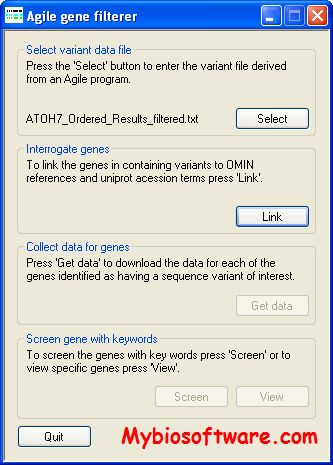
:: SCREENSHOTS
N/a
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Yu C, Zhao Y, Zhao C, Ma H, Wang G.
DiagAF: A More Accurate and Efficient Pre-Alignment Filter for Sequence Alignment.
IEEE/ACM Trans Comput Biol Bioinform. 2021 Nov 17;PP. doi: 10.1109/TCBB.2021.3127879. Epub ahead of print. PMID: 34780330.