VarioWatch 20171116
:: DESCRIPTION
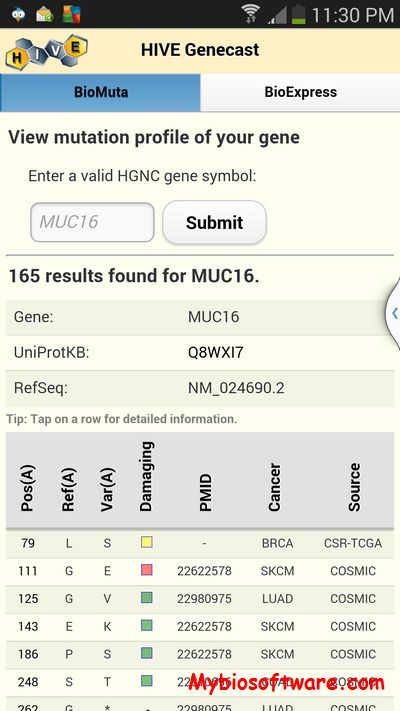
VarioWatch is a functional variant and disease gene mining browser for association study
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
Nucleic Acids Res. 2012 Jul;40(Web Server issue):W76-81. doi: 10.1093/nar/gks397. Epub 2012 May 22.
VarioWatch: providing large-scale and comprehensive annotations on human genomic variants in the next generation sequencing era.
Cheng YC1, Hsiao FC, Yeh EC, Lin WJ, Tang CY, Tseng HC, Wu HT, Liu CK, Chen CC, Chen YT, Yao A.