SNPYGoat 1.0
:: DESCRIPTION
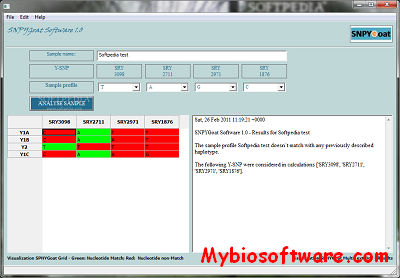
SNPYGoat Software allows users of the SNPYGoat multiplex system to rapidly identify several goat Y-chromosomal haplotypes Y1A, Y1B, Y1C and Y2 by automatically comparing the obtained profile with a reference database.
::DEVELOPER
:: SCREENSHOTS
: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Pereira F, Carneiro J, Soares P, Maciel S, Nejmeddine F, Lenstra JA, Gusm?o L, Amorim A
A multiplex primer extension assay for the rapid identification of paternal lineages in domestic goat (Capra hircus): laying the foundations for a detailed caprine Y chromosome phylogeny
Molecular Phylogenetics and Evolution. 2008. 49:663-668