PexSPAM 1.2
:: DESCRIPTION
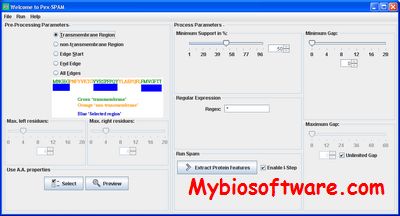
PexSPAM is a Java standalone program that I wrote for protein sequence feature extraction. PexSPAM was originally designed to be a “feature factory” for secondary structure classification problem in integral membrane proteins. PexSPAM extends the SPAM (Ayres et al, 2002) method by incorporating gap and regular expression constraints into mining procedure.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / MacOs / Linux / Unix
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Ho J, Lukov L, Chawla S (2005)
Sequential Pattern Mining with Constraints on Large Protein Databases.
In Chakrabarti S, Sudarshan S, Radha Krishnan P (Eds) Proceedings of the 12th International Conference on Management of Data (COMAD 2005b), 89-100.