ProteinVis 2.1.6
:: DESCRIPTION
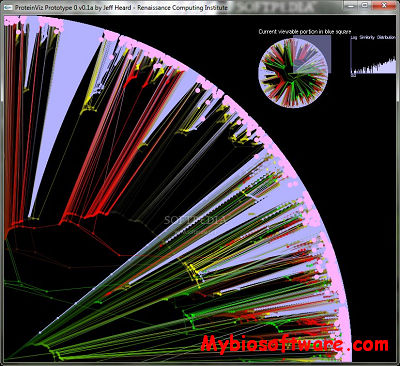
ProteinVis is a tree viewer for hierarchical clusterings of proteins in the human Genome.Many genomic and proteomic analyses generate as a result a tree of genes or proteins. These trees are often large (containing tens of thousands of nodes and edges), and need a visualization tool to fully display all the information contained in the tree. Clustering analysis can be performed on these trees to obtain clusters of proteins, and we need an efficient way to visualize the clustering results. We present a novel tree visualization tool to help with such analyses.
::DEVELOPER
Jefferson Robert Heard ; Xiaojun Guan, and a professor of biology at UNC Chapel Hill, Dr William Kaufman.
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux
:: DOWNLOAD
 ProteinVis for win ; for Linux
ProteinVis for win ; for Linux
:: MORE INFORMATION
Citation
Jeff Heard1,, William Kaufmann and Xiaojun Guan1,
A novel method for large tree visualization
Bioinformatics (2009) 25 (4): 557-558.