pGQL
:: DESCRIPTION
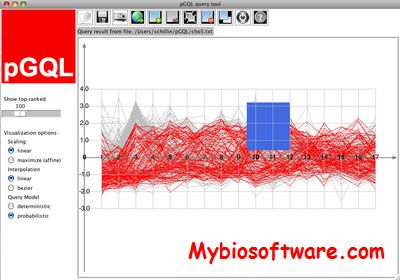
pGQL (probabilistic Graphical Query Language) is a software tool in particular for analyzing gene expression time courses. It allows its user to interactively define linear HMM queries on time course data using rectangular graphical widgets called probabilistic time boxes. The analysis is fully interactive and the graphical display shows the time courses along with the graphical query. The results can be submitted to gPROF directly from pGQL.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/ MacOsX/ Windows
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation
BioData Min. 2011 Apr 18;4:9. doi: 10.1186/1756-0381-4-9.
pGQL: A probabilistic graphical query language for gene expression time courses.
Schilling R, Costa IG, Schliep A.