SMRT Analysis 2.3.0
:: DESCRIPTION
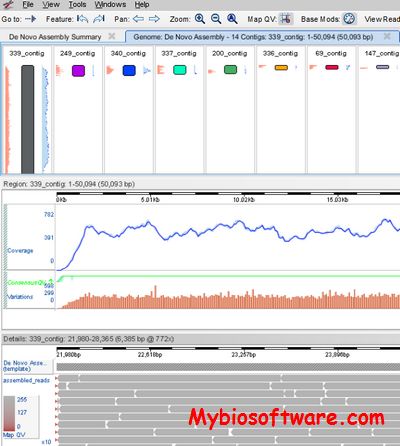
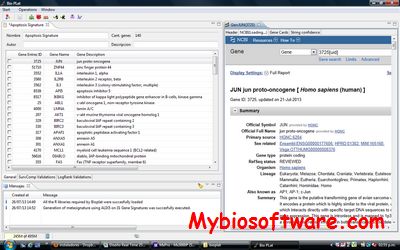
SMRT Analysis is a bioinformatics software suite for analyzing single molecule, real-time DNA sequencing data from Pacific Biosciences.
::DEVELOPER
Pacific Biosciences of California, Inc
:: SCREENSHOTS
:: REQUIREMENTS
- Linux
- perl
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Genome Integr. 2011 Dec 20;2:10. doi: 10.1186/2041-9414-2-10.
Direct detection and sequencing of damaged DNA bases.
Clark TA, Spittle KE, Turner SW, Korlach J.