expVIP 20180914
:: DESCRIPTION
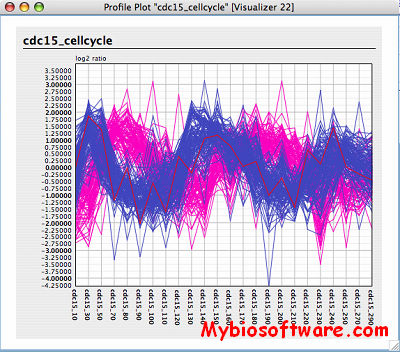
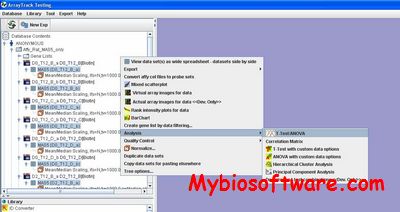
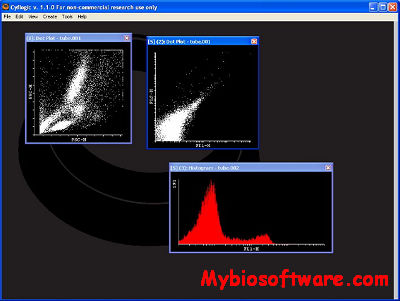
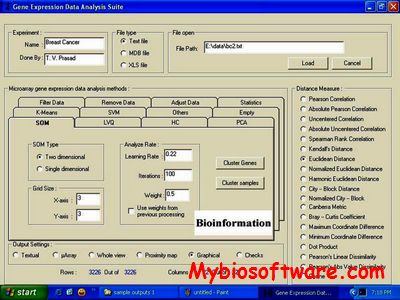
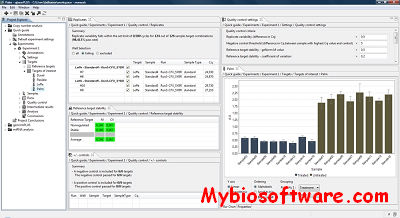
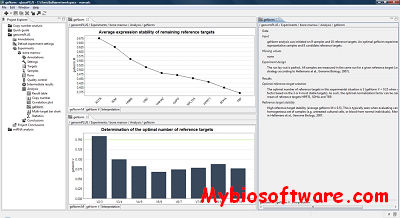
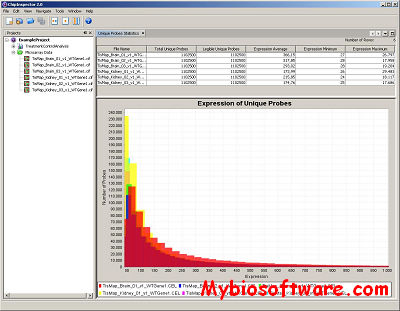
ExpVIP (expression visualization and integration platfor) is a tool that enables the integration of different RNA-Seq studies by analysing the data uniformly across experiments and provides a web interface to query the data.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
expVIP: a Customizable RNA-seq Data Analysis and Visualization Platform.
Borrill P, Ramirez-Gonzalez R, Uauy C.
Plant Physiol. 2016 Apr;170(4):2172-86. doi: 10.1104/pp.15.01667.