qPCR-DAMS 1.2
:: DESCRIPTION
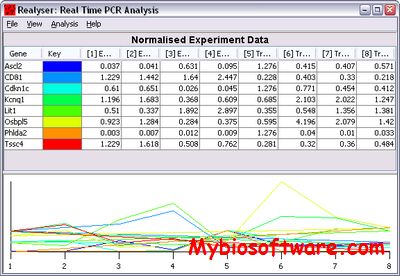
qPCR-DAMS is a database management system implemented on MS Access 2003 and Visual Basic. It is designed to analyze, manage, and store relative and absolute quantitative real-time PCR data. The system consists of 5 blocks: three blocks (Gene, Plate, and Experiment) for inputting, storing and describing raw data, and two blocks (View data and Process Data) for checking, evaluating, and processing data. Users are allowed to choose among four basic outputs: (I) Ratio relative quantification, (II) Absolute level, (III) Normalized absolute expression, and (IV) Ratio absolute quantification, and two advanced options: (V) Multiple reference relative quantification and (VI) Multiple references absolute quantification, within single software package. The coefficient of variation is monitored at each step during data processing and the accuracy is further improved by an easy data tracking and display system. In summary, qPCR-DAMS is a handy novel tool for real-time PCR users.
::DEVELOPER
Nili Jin , Keyu He and Lin Liu
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows
- Microsoft Access 2000 or above
:: DOWNLOAD
 qPCR-DAMS
qPCR-DAMS
:: MORE INFORMATION
Citation
Nili Jin , Keyu He and Lin Liu
qPCR-DAMS: a database tool to analyze, manage, and store both relative and absolute quantitative real-time PCR data
Physiological Genomics 25:525-527 (2006)