CARMAweb 1.3
:: DESCRIPTION
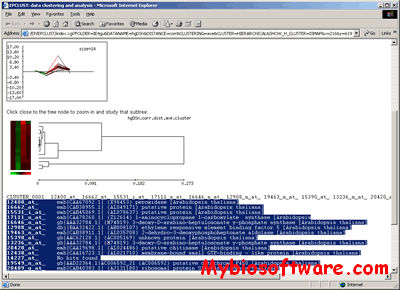
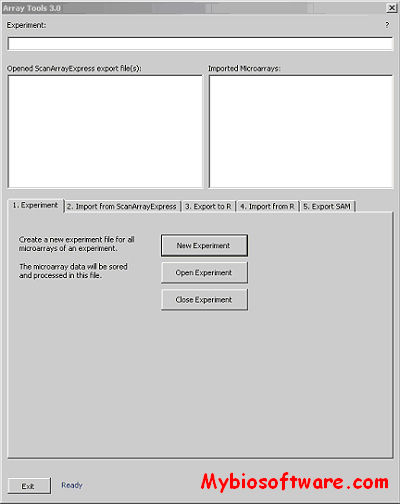
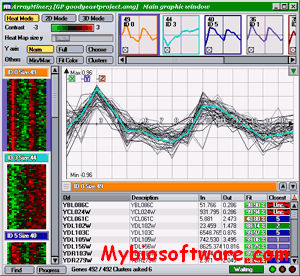
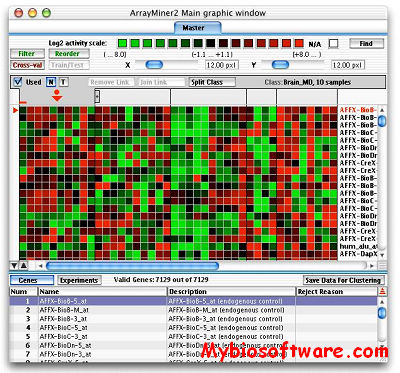
CARMAweb (Comprehensive R-based Microarray Analysis web service) is a web application designed for the analysis of microarray data. CARMAweb performs data preprocessing (background correction, quality control and normalization), detection of differentially expressed genes, cluster analysis, dimension reduction and visualization, classification, and Gene Ontology-term analysis. This web application accepts raw data from a variety of imaging software tools for the most widely used microarray platforms: Affymetrix GeneChips, spotted two-color microarrays and Applied Biosystems (ABI) microarrays.
::DEVELOPER
Genomics & Bioinformatics Graz, Graz University of Technology
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- R Package
- Bioconductor
:: DOWNLOAD
:: MORE INFORMATION
Citation
Rainer J., Sanchez-Cabo F., Stocker G., Sturn A. and Trajanoski Z.
CARMAweb: comprehensive R- and Bioconductor-based web service for microarray data analysis
Nucleic Acids Research, 2006 vol 34(Web Server Issue):W498-503.