JSim 2.15
:: DESCRIPTION
JSim is a Java-based simulation system for building and analyzing quantitative numeric models. JSim’s primary focus has been in physiology and bio-medicine, however its computation engine is quite general and applicable to a wide range of scientific domains.
::DEVELOPER
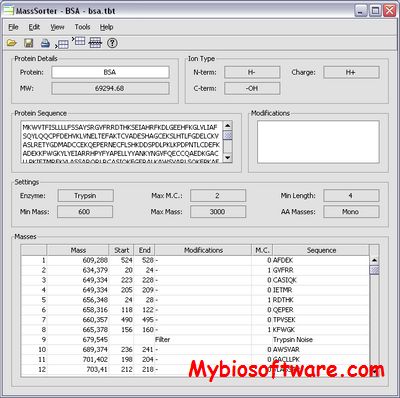
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux / Mac OsX
- JAVA
:: DOWNLOAD
:: MORE INFORMATION
Citation
JSim, an open-source modeling system for data analysis.
Butterworth E, Jardine BE, Raymond GM, Neal ML, Bassingthwaighte JB.
F1000Res. 2013 Dec 30;2:288. doi: 10.12688/f1000research.2-288.v1.