OFF-TARGET PIPELINE 1.5.4.2
:: DESCRIPTION
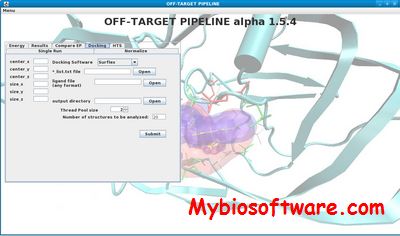
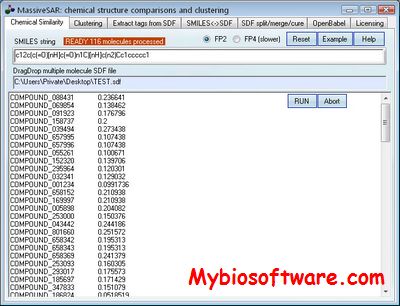
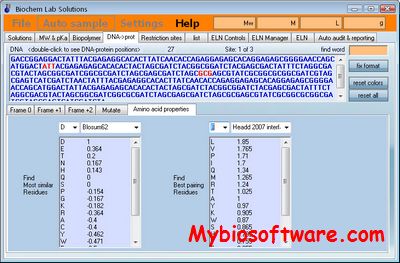
OFF-TARGET PIPELINE is a platform intended to carry out a recently introduced chemical systems biology approach for secondary target identification, but may also be useful to other applications in bioinformatics and drug discovery. Researchers can predict and compare protein functional sites, conduct structure-based docking, compute binding energy and compare electrostatic potential distribution within cavities. It also supports normalization of docking scores, information retrieval from PDB, and parsing of output files. All the afore-mentioned procedures are parallelized with significant speed gains and labor savings.
::DEVELOPER
Thomas Evangelidis (tevang3@gmail.com)
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
References:
L. Xie, J. Wang and P.E. Bourne 2007
“In Silico Elucidation of the Molecular Mechanism Defining the Adverse Effect of Selective Esterogen Receptor Modulators“.
PLoS Comp. Biol., 3(11):e217