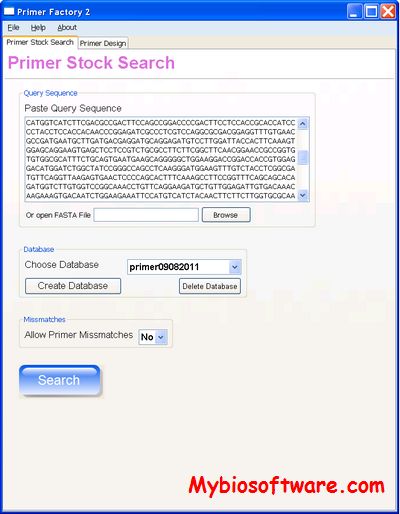
morePhyML 1.14
:: DESCRIPTION
morePhyML has been implemented to improve ML tree space exploration with PhyML. morePhyML allows inferring more accurate phylogenetic trees than several other recently developed ML tree inference softwares in many cases.
::DEVELOPER
Alexis Criscuolo (alexis.criscuolo@pasteur.fr)
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Criscuolo A. (2011)
morePhyML: improving the phylogenetic tree space exploration with PhyML 3.
Mol Phylogenet Evol. 61(3):944-8.