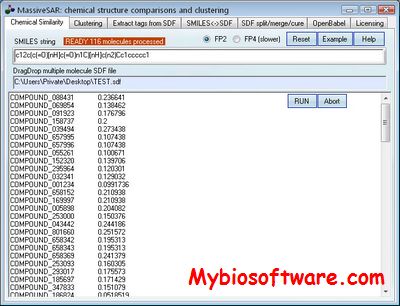
GMASS
:: DESCRIPTION
The GMASS score is a novel measure for representing structural similarity between two assemblies. It represents the structural similarity of a pair of assemblies based on the length and number of similar genomic regions defined as consensus segment blocks (CSBs) in the assemblies.
::DEVELOPER
Bioinformatics Laboratory, Konkuk University
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- Perl
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2019 Mar 18;20(1):147. doi: 10.1186/s12859-019-2710-z.
GMASS: a novel measure for genome assembly structural similarity.
Kwon D, Lee J, Kim J.