CN3D 4.3.1
:: DESCRIPTION
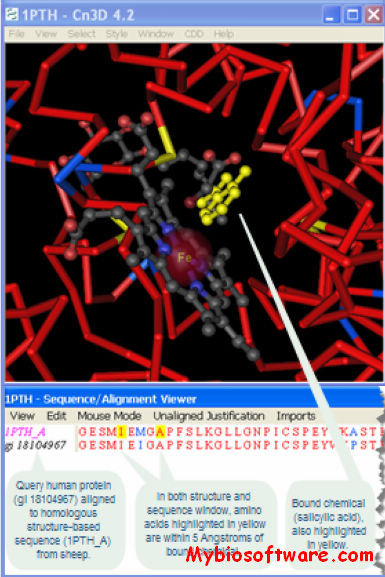
Cn3D is a helper application for your web browser that allows you to view 3-dimensional structures from NCBI’s Entrez retrieval service. Cn3D simultaneously displays structure, sequence, and alignment, and now has powerful annotation and alignment editing features.
What sets Cn3D apart from other software is its ability to correlate structure and sequence information: for example, a scientist can quickly find the residues in a crystal structure that correspond to known disease mutations, or conserved active site residues from a family of sequence homologs. Cn3D displays structure-structure alignments along with their structure-based sequence alignments, to emphasize what regions of a group of related proteins are most conserved in structure and sequence.
Cn3D reads only data files from the MMDB database, not PDB formatted records.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Macintosh/Unix/Linux
:: DOWNLOAD
:: MORE INFORMATION
Below is a complete list (with links into Entrez) of all literature cited in the preparation of this tutorial.
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell 2000 Feb 18; 100(4):387-90. (Entrez)
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 1999 Oct 29; 99(3):323-34. (Entrez)
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997 May; 16(1):64-7. (Entrez)