ACES
:: DESCRIPTION
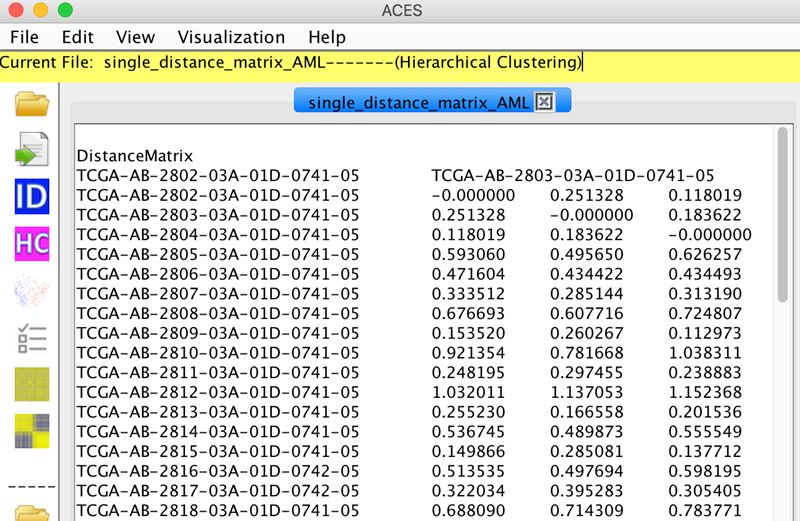
ACES is a machine learning toolbox for clustering analysis and visualization of both biological data and other types data. Given the biological data or their distance/probability matrix, ACES can automatically extract the features of each identity and cluster them by various widely used clustering algorithms.
::DEVELOPER
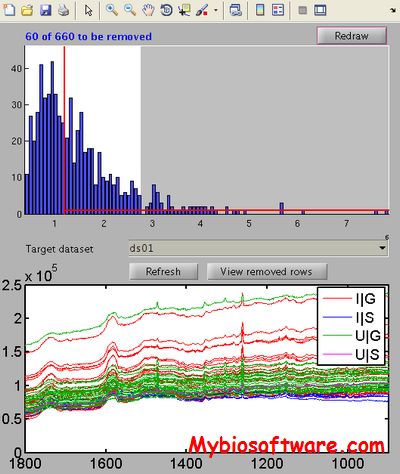
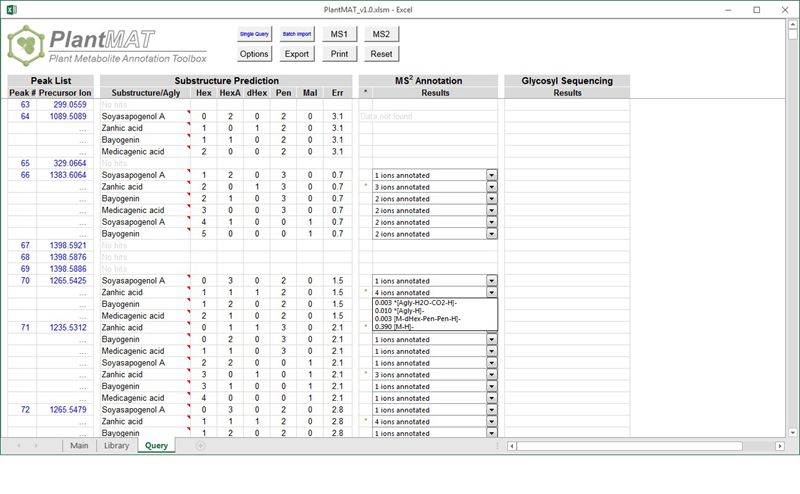
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Gao J, Sundström G, Moghadam BT, Zamani N, Grabherr MG.
ACES: a machine learning toolbox for clustering analysis and visualization.
BMC Genomics. 2018 Dec 27;19(1):964. doi: 10.1186/s12864-018-5300-y. PMID: 30587115; PMCID: PMC6307290.