CNApy 1.0.6
:: DESCRIPTION
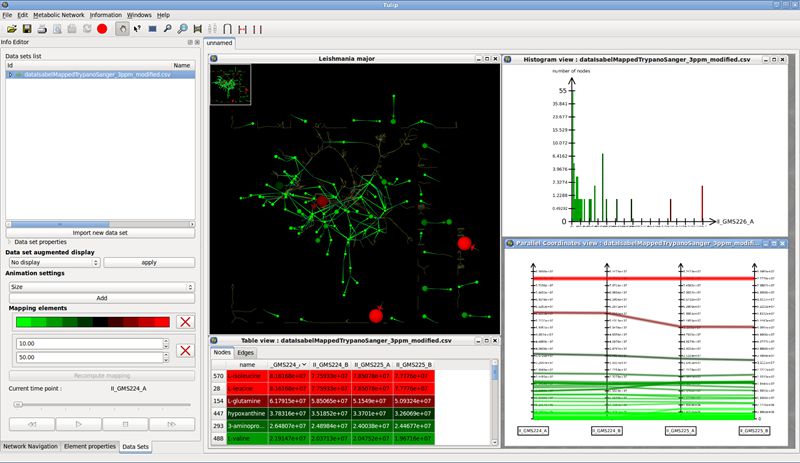
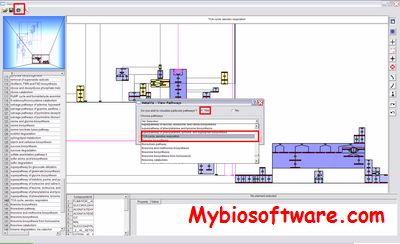
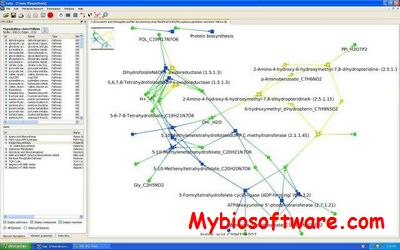
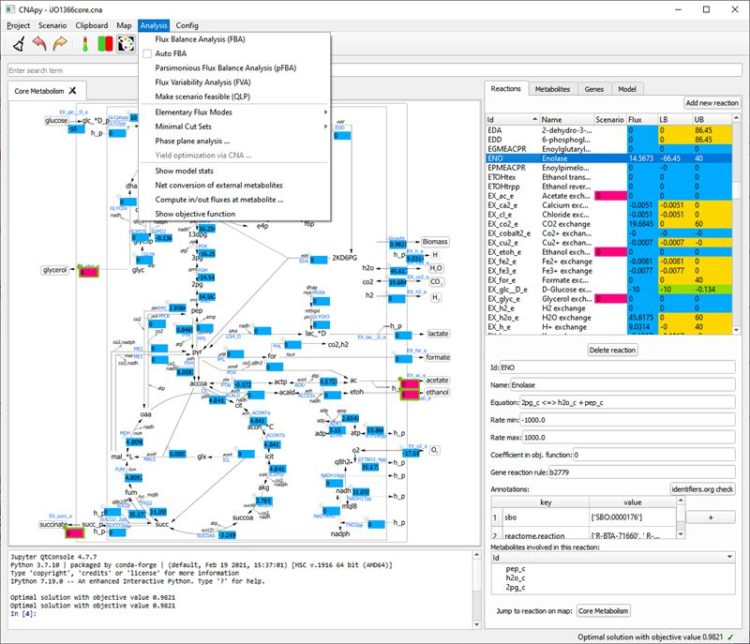
CNApy is an open source cross-platform desktop application written in Python, which offers a state-of-the-art graphical front-end for the intuitive analysis of metabolic networks with COBRA methods.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Thiele S, von Kamp A, Bekiaris PS, Schneider P, Klamt S.
CNApy: a CellNetAnalyzer GUI in Python for Analyzing and Designing Metabolic Networks.
Bioinformatics. 2021 Dec 8:btab828. doi: 10.1093/bioinformatics/btab828. Epub ahead of print. PMID: 34878104.