dasty 3.0.0.9
:: DESCRIPTION
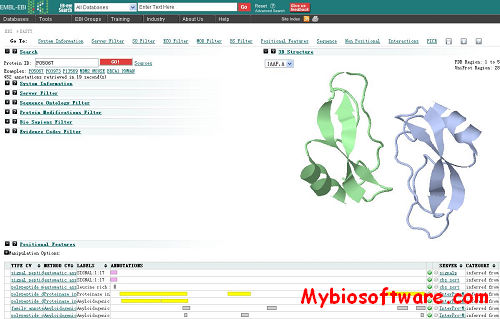
Dasty3 is a web client for visualizing protein sequence feature information using DAS. Through the DAS registry the client establishes connections to a DAS reference server to retrieve sequence information and to one or more DAS annotation servers to retrieve feature annotations. It merges the collected data from all these servers and provides the user with a unified, aesthetically pleasing, effective view of the sequence-annotated features. Dasty3 uses AJAX to deliver highly interactive graphical functionality in a web browser by executing multiple asynchronous DAS requests.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Dasty2, an Ajax protein DAS client.
Jimenez RC, Quinn AF, Garcia A, Labarga A, O’Neill K, Martinez F, Salazar GA, Hermjakob H
(15-Sep-2008) Bioinformatics (Oxford, England), 24 (18) :2119-21