imappingfamily
:: DESCRIPTION
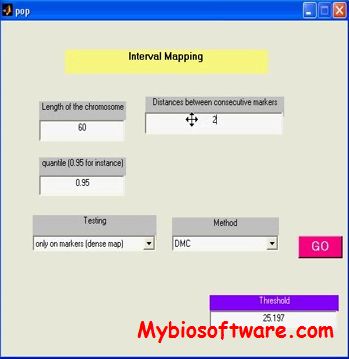
imappingfamily is a package with a graphical interface regrouping the different methods presented in the article submitted “Quantitative Trait Locus Detection in a population with family structure”.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/windows / MacOsX
- Matlab
:: DOWNLOAD
:: MORE INFORMATION
Citation
Rabier C-E, Azais J-M, Elsen J-M, Delmas C.
Quantitative Trait Locus Detection in a population with family structure
(submitted)