Cri-Map 2.4
:: DESCRIPTION
CRI-MAP‘s purpose is to allow rapid, largely automated construction of multilocus linkage maps (and facilitate the attendant tasks of assessing support relative to alternative locus orders, generating LOD tables, and detecting data errors). Although originally designed to handle codominant loci (e.g. RFLPs) scored on pedigrees “without missing individuals”, such as CEPH or nuclear families, it can now (with some caveats described below) be used on general pedigrees, and some disease loci.
::DEVELOPER
Matise Laboratory of Computational Genetics.
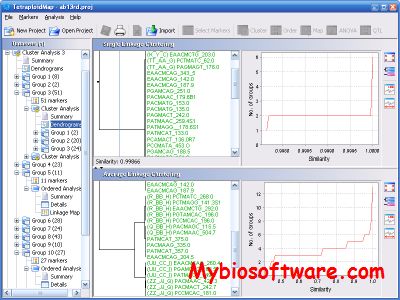
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows wity Cygwin /MacOsX
- GCC
:: MORE INFORMATION
Citation
Hum Hered. 1995 Mar-Apr;45(2):103-16.
Parallel computation of genetic likelihoods using CRI-MAP, PVM, and a network of distributed workstations.
Matise TC, Schroeder MD, Chiarulli DM, Weeks DE.