Pedigree-Draw 6
:: DESCRIPTION
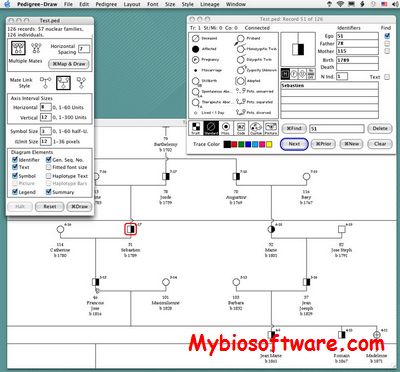
Pedigree-Draw is a software that provides for creation, editing and drawing of pedigrees, or family trees, of human or non-human extended family lineages. It was developed originally for use in analyzing population data gathered for biomedical and genetic research. Although not designed as a genealogical database per se, the program can be used to collect and edit biographical data found in the course of family history research, as well as pictures of family members, and display or print those data in hierarchical diagrams suitable for publication. Output may also be saved in Portable Document Format (PDF) for distribution via the Internet, or posting on websites.
::DEVELOPER
Jurek Software
:: SCREENSHOTS
:: REQUIREMENTS
:: DOWNLOAD
 Pedigree-Draw
Pedigree-Draw
:: MORE INFORMATION

