ExactSearch
:: DESCRIPTION
ExactSearch is a web tool which enables plant biologists to search for DNA motifs in the proximal promoters, and 3′ untranslated regions of all genes from 50 genome-sequenced plant species.
::DEVELOPER
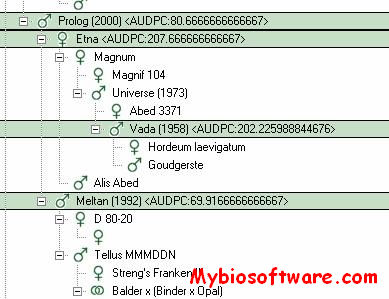
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
ExactSearch: a web-based plant motif search tool.
Gunasekara C, Subramanian A, Avvari JV, Li B, Chen S, Wei H.
Plant Methods. 2016 Apr 28;12:26. doi: 10.1186/s13007-016-0126-6.