PlantMAT v1.0
:: DESCRIPTION
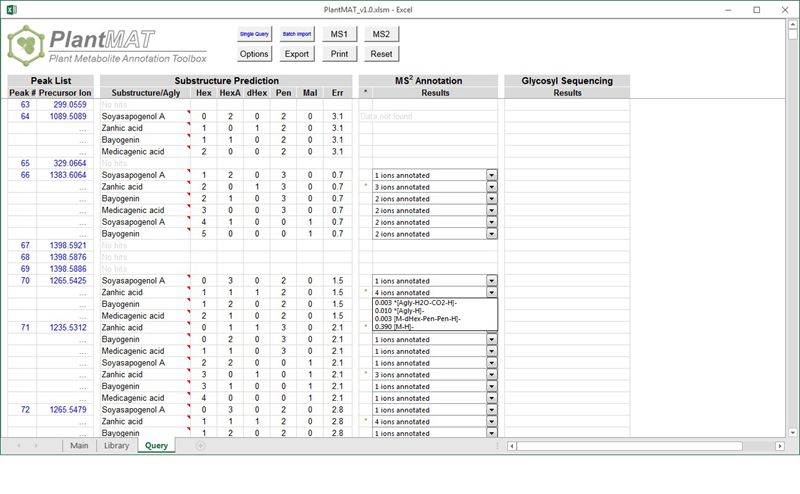
PlantMAT is a computational tool combining library search and combinatorial enumeration to facilitate the structural identification of plant specialized metabolites, e.g., saponins and flavonoid glycosides.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
- Excel
:: DOWNLOAD
:: MORE INFORMATION
Citation
Qiu F, Fine DD, Wherritt DJ, Lei Z, Sumner LW.
PlantMAT: A Metabolomics Tool for Predicting the Specialized Metabolic Potential of a System and for Large-Scale Metabolite Identifications.
Anal Chem. 2016 Dec 6;88(23):11373-11383. doi: 10.1021/acs.analchem.6b00906. Epub 2016 Nov 23. PMID: 27934098.