PESCADOR
:: DESCRIPTION
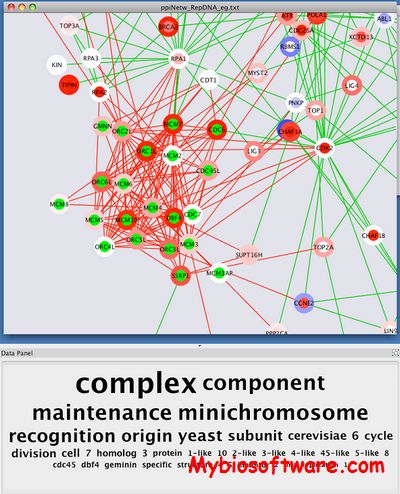
PESCADOR (Platform for Exploration of Significant Concepts AssociateD to co-Occurrences Relationships) is a web tool that extracts a network of interactions from a set of PubMed abstracts given by a user, and allows filtering the interaction network according to user-defined concepts.
::DEVELOPER
Computational Biology and Data Mining (CBDM) Group
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Server
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
BMC Bioinformatics. 2011 Nov 9;12:435. doi: 10.1186/1471-2105-12-435.
PESCADOR, a web-based tool to assist text-mining of biointeractions extracted from PubMed queries.
Barbosa-Silva A1, Fontaine JF, Donnard ER, Stussi F, Ortega JM, Andrade-Navarro MA.